Research
Single Cell Analysis of Adipose Tissue from Mouse and Human
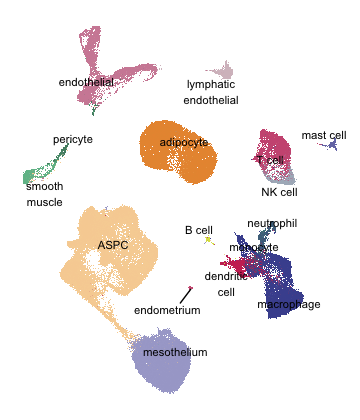
Adipose tissue is complex and heterogeneous; in the normal lean state, white fat is composed of roughly 50% adipocytes and 50% other cell types, such as endothelial cells, preadipocytes, fibroblasts, and immune cells. In obesity, the ratio of nonadipocytes: adipocytes can rise to as high as 10:1! Understanding adipose tissue biology thus requires a detailed accounting of the constituent cell types and how they change according to factors like depot type, sex, age, nutritional status, or ambient temperature. Unfortunately, single whole cell approaches are not suitable for capturing adipocytes, which are too large and fragile to withstand the shear forces associated with microfluidics. Accordingly, we have optimized a single nucleus (snRNA-seq) approach for adipose tissue, and have utilized it to generate an Adipose Atlas (v1.0) (PMID: 35296864). This atlas compares white adipose tissue across species, sexes, depots, and body mass, and can be viewed on the Broad Single Cell Portal (link). This work enabled us to identify new subpopulations of adipocytes, find associations between specific adipose cell types and metabolic disease traits, and develop an initial roadmap of cell-cell communication within the adipose niche. Ongoing work is focused on profiling additional cells from different depots and disease states, and in better characterizing adipocyte heterogeneity.
Deconvoluting the Genetic Basis of Human Insulin Resistance

Why do human beings get insulin resistance and type 2 diabetes (T2D)? We know a lot about the signaling and transcriptional networks that promote or oppose insulin action in cultured cells and in mouse models, but are these the most relevant pathways in humans? In order to answer this, we can turn to studies of human genetics. GWAS analysis of human T2D has now implicated over 240 loci with roughly 500 independent association signals. The vast majority of these signals are located in noncoding regions of the genome. In virtually all cases for which it has been scrutinized, the mechanism of action of noncoding variants is to alter transcription factor binding and thus expression of key target genes. In this light, diseases like T2D and IR can be thought of as ‘enhancer-opathies’. In collaboration with Linus Tsai (BIDMC), Melina Claussnitzer (Broad), and others, we are using high-throughput CRISPR-based screening techniques to identify genes and enhancers that contribute to insulin resistance in human adipocytes. We predict which variants are most likely to be causal in disease pathogenesis, and then validate these predictions using variant switching. We are also identifying the upstream transcription factors whose binding is compromised by the presence of the variant.
Deciphering the Roles of Interferon Regulatory Factors in Metabolism
It has been known for some time that obesity is accompanied by low-grade inflammation of adipose tissue and other metabolic organs, and that inflammation helps drive some of the adverse sequelae of excess fat, such as insulin resistance, suppression of thermogenesis, and dyslipidemia. We have been interested in identifying the transcription factors and pathways that mediate these effects, and have focused on members of the interferon regulatory factor (IRF) family. The nine mammalian IRFs have been implicated in almost every aspect of immunity, from macrophage formation and lymphopoieisis to T cell activation, but we were the first to define a role for IRFs in adipose biology and metabolic function. Most of our work has focused on IRF3 and IRF4, which are highly expressed in adipocytes and which we have shown are regulators of adipogenesis (PMID: 18177728). IRF3 is a pro-inflammatory factor that promotes insulin resistance and obesity on a high fat diet, related at least in part to its anti-thermogenic properties (PMID: 27400129, 33571167). IRF4 on the other hand, is pro-thermogenic, and is the natural DNA binding partner of PGC-1a in brown fat (PMID: 24995979). We have also shown that IRF4 in brown fat regulates the expression of myostatin, which in turn controls skeletal muscle function (PMID: 30078553). Thus, animals lacking IRF4 in BAT have an unusual form of myopathy and diminished exercise capacity, which is pretty cool. We are currently pursuing several lines of investigation related to the actions of IRFs, including: (a) What are the tissue-specific roles of IRF3 in macrophages vs. adipocytes?; (b) Is IRF3 required for the development of fatty liver; (c) What are the critical downstream target genes of IRF3 and IRF4 in fat and liver, and how do they impact upon metabolic function in those tissues?; (d) What are the key post-translational modifications and interacting partners of IRFs in fat and liver?; and (e) What other IRFs are important metabolic regulators in fat and other tissues?
Elucidating Adipose-Lymphatic Interactions
The lymphatic system comprises a series of blind-ended capillaries and collecting ducts which empty into the blood circulation; its purpose is to return extravasated fluid, solutes, and immune cells to the circulation. Like most organs, adipose tissue has a lymphatic vasculature, but there is little insight into whether or how it contributes to the function of fat (for our review on the subject, see PMID: 34718498). Furthermore, chronic lymphedema is a serious and prevalent condition in which defective lymphatic drainage (such as after surgery) leads to abnormal accumulation of adipose tissue. We are using a variety of approaches to dissect out potential cross-talk between the lymphatic system and adipose tissue in normal and abnormal conditions. As one example, we have discovered that lymphatic endothelial cells make the neuropeptide neurotensin (NTS), which acts to inhibit adipose thermogenesis. NTS expression and secretion are negatively regulated by sympathetic release of norepinephrine, providing an alternative mechanism by which catecholamines induce heat production in fat (PMID: 34038712). Ongoing studies are focused on cellular abnormalities in chronic lymphedema and the role of adipokines (such as leptin) on lymphatic function.
