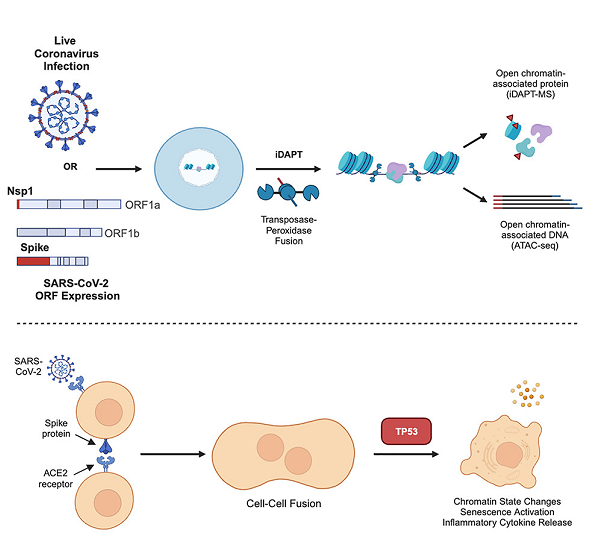
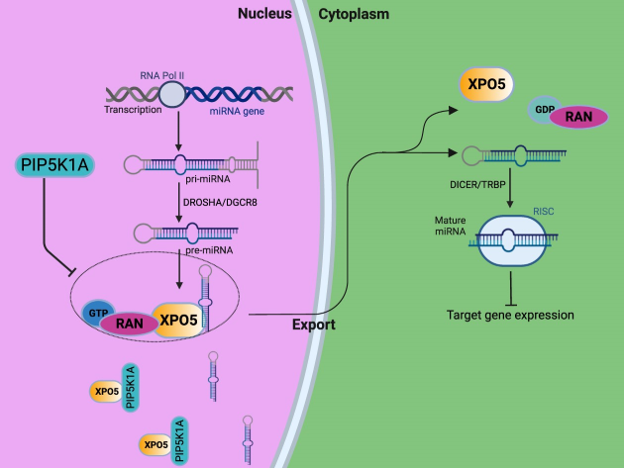
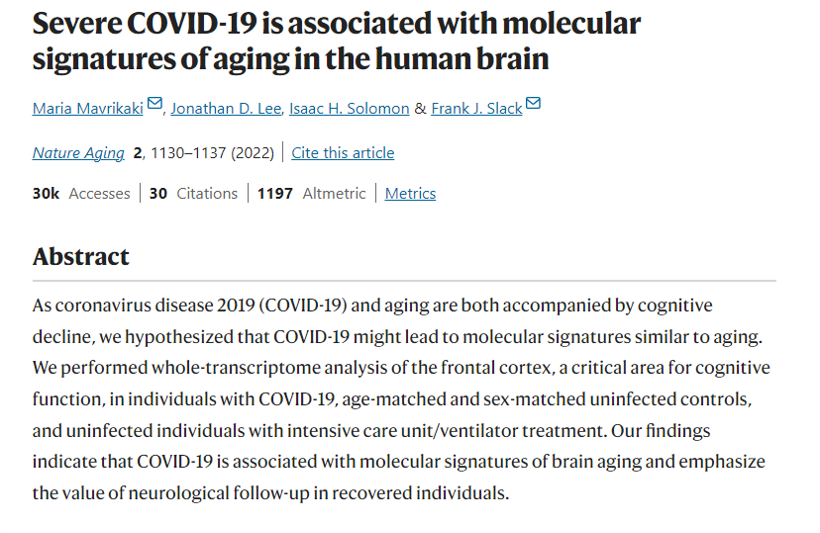
Welcome to the Slack Lab!
The past decade has brought about a paradigm shift in how the scientific community views gene regulation, and we are thrilled to be at the vanguard of this biological revolution. Non-coding regulatory RNAs direct such diverse processes as gene silencing, transcriptional and translational control, imprinting and dosage compensation. These discoveries have electrified the biological community as we try to understand the extent of the “RNA world” and to harness these new molecules in medicine [1]. One class of regulatory RNAs leading the charge in this revolution are the microRNAs (miRNAs). MiRNAs are recently discovered regulatory molecules that control a large variety of biological processes including development, metabolism and aging [2-4]. The Slack lab co-discovered the second known miRNA and the first human miRNA [5, 6]. Since then, hundreds of miRNAs have been identified in worm, fly, and mammalian genomes, However, the biological roles of only a small fraction of these miRNAs have been characterized. Members of the Slack lab have continued to push the boundaries of this exciting field by, for example, discovering roles for microRNAs in cancer and aging [7, 8] and in translating these discoveries to the clinic as potential cures and biomarkers for cancer and aging [9-12].
Regards,
Frank Slack, PhD
Shields Warren Mallinckrodt Professor of Medical Research
Harvard Medical School
Director of the Harvard Initiative for RNA Medicine
Director of the Cancer Research Institute for RNA Medicine
Departments of Pathology and Medicine
Beth Israel Deaconess Medical Center
- Slack, F.J., Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biol, 2006. 7(9): p. 328.
- Banerjee, D. and F. Slack, Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays, 2002. 24(2): p. 119-29.
- Grosshans, H. and F.J. Slack, Micro-RNAs: small is plentiful. J Cell Biol, 2002. 156(1): p. 17-22.
- Niwa, R. and F.J. Slack, The evolution of animal microRNA function. Curr Opin Genet Dev, 2007. 17(2): p. 145-50.
- Pasquinelli, A.E., et al., Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 2000. 408(6808): p. 86-9.
- Reinhart, B.J., et al., The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 2000. 403(6772): p. 901-6.
- Boehm, M. and F. Slack, A developmental timing microRNA and its target regulate life span in C. elegans. Science, 2005. 310(5756): p. 1954-7.
- Johnson, S.M., et al., RAS is regulated by the let-7 microRNA family. Cell, 2005. 120(5): p. 635-47.
- Esquela-Kerscher, A., et al., The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle, 2008. 7(6): p. 759-64.
- Medina, P.P., M. Nolde, and F.J. Slack, OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 2010. 467(7311): p. 86-90.
- Trang, P., et al., Regression of murine lung tumors by the let-7 microRNA. Oncogene, 2010. 29(11): p. 1580-7.
- Trang, P., et al., Systemic Delivery of Tumor Suppressor microRNA Mimics Using a Neutral Lipid Emulsion Inhibits Lung Tumors in Mice. Mol Ther, 2011. 19(6): p. 1116-22.