Research
Polygenic and Omics Scores
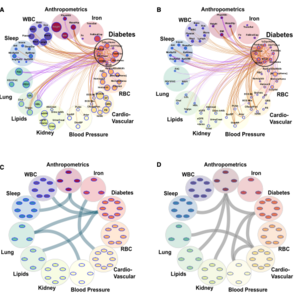
We develop and apply cutting-edge statistical methods for genetic and omics association analyses. Our research projects include:
(1) developing polygenic risk score (PRS) methods to analyze sleep-related phenotypes (e.g., obstructive sleep apnea PRS, short sleep PRS) and cardiovascular-related diseases (e.g., hypertension PRS, blood pressure PRS) in multi-ethnic populations.
(2) applying the developed PRSs to conduct PRS-outcome association analyses and identify population-specific effect estimates.
(3) constructing proteomics and metabolomics risk scores using penalized regression and conducting association analyses to estimate the effect sizes between omics scores and the phenotypes of interest. We use polygenic and omics scores to understand how phenotypes are related to each other, and incorporate them in prediction models.
Please see below for the research being completed in Sofer's Lab.
Using polygenic risk scores to study health
One of the main interests of our lab includes leveraging information on the individual’s genetic predisposition to traits and diseases, such as insomnia or hypertension, to better understand genetic architecture of the trait or disease and the overall role of genetics in our health. We measure the genetic burden of traits and diseases for an individual using the polygenic risk score, or PRS, for that trait or disease where the dosage of risk alleles in genome is weighted by their respective effect size and summed across the genome. We then can stratify individuals into low and high-risk groups based on the lower and higher PRS values respectively within the population. This can help allocate healthcare resources more efficiently and design targeted public health interventions. Furthermore, we can study the cumulative effects of individual’s genetic predisposition to a certain disease (i.e. PRS for systolic and diastolic blood pressure) and their lifestyle (smoking and sleeping habits) on their health, by estimating the association between their PRS, demographic and lifestyle data with health-related outcomes (i.e. hypertension).
Prediction using genetics and omics
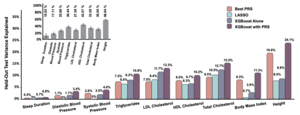
Our lab is in construction of Machine Learning-based polygenic prediction models for estimating individual’s likelihood of acquiring poor cardiovascular traits (i.e. hypertension) in the future according to their current lifestyle paired with their genetic predisposition to the trait. In addition to the individual’s genetics, we are using their omics measures, such as levels of metabolites and proteins in blood.
Omics measures have been increasingly used to capture individual’s health status and risks. Omics have the advantage of being modifiable via positive changes in lifestyle and clinical intervention, compared to individual’s genetics which are fixed at birth. We integrate omics biomarkers as predictors for developing blood pressure phenotypes based on the individual’s current state. We hypothesize that the effects of genetics together with lifestyle, clinical, and other environmental exposures on blood pressure are likely mediated via omics measures. Metabolomics and proteomics are closer to the phenotype level, in comparison to genetics, and thus may be particularly useful as biomarkers.
Model prediction based on clinical and current lifestyle data together with blood pressure PRS, proteome and metabolites scores quantify relative effects of lifestyle, genetics and omics on the likelihood of acquiring blood pressure phenotypes. All together this may allow for identifying modifiable pathways to cardiovascular health.
Study of determinants of sex differences
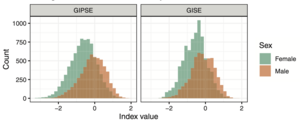
Sex differences are everywhere. Obstructive sleep apnea is more common in men. Insomnia is more common in women. Alzheimer’s disease is more common in women. And so on. But why? The better we understand patterns of sex differences and the better we understand reasons behind sex differences, the better we will be in personalizing medicine for men and women. We are studying differences in both genetic and non-genetic determinants of health and disease outcomes between groups of individuals categorized by sex groups (and we are currently focusing only on male and female sex). For example, our analyses demonstrate that genetic determinants of obstructive sleep apnea are associated with increased risk of hypertension primarily in women, but not in men (genetic determinants). We also see that differences in occupation and earning relate to differences in insomnia between men and women (non-genetic determinant). For example, in the figure, we developed "gendered indices" summarizing sociodemographic variables and overall having different distributions between male and female individuals. In addition to performing sophisticated analyses to study sex differences, we are also currently developing statistical methods to address them. Stay tuned!
Statistical methods for genetic association analyses
We develop new statistical methods for genetic association analyses. Examples include rare variant association tests, methods for estimating heritability and genetic correlations, and to accurately estimate uncertainly (e.g. develop well-calibrated confidence intervals) methods for calibrating polygenic risk scores to admixed populations. These methods are used to understand genetic determinants of complex phenotypes and omics phenotypes. For example, to understand genetic determinants of protein levels and genetic regulation of protein networks. Ultimately, our goal is to improve our understanding of health and disease in diverse and understudied populations.
