Research
Transcriptional Regulation of Human Leukocyte Antigen (HLA) Molecules
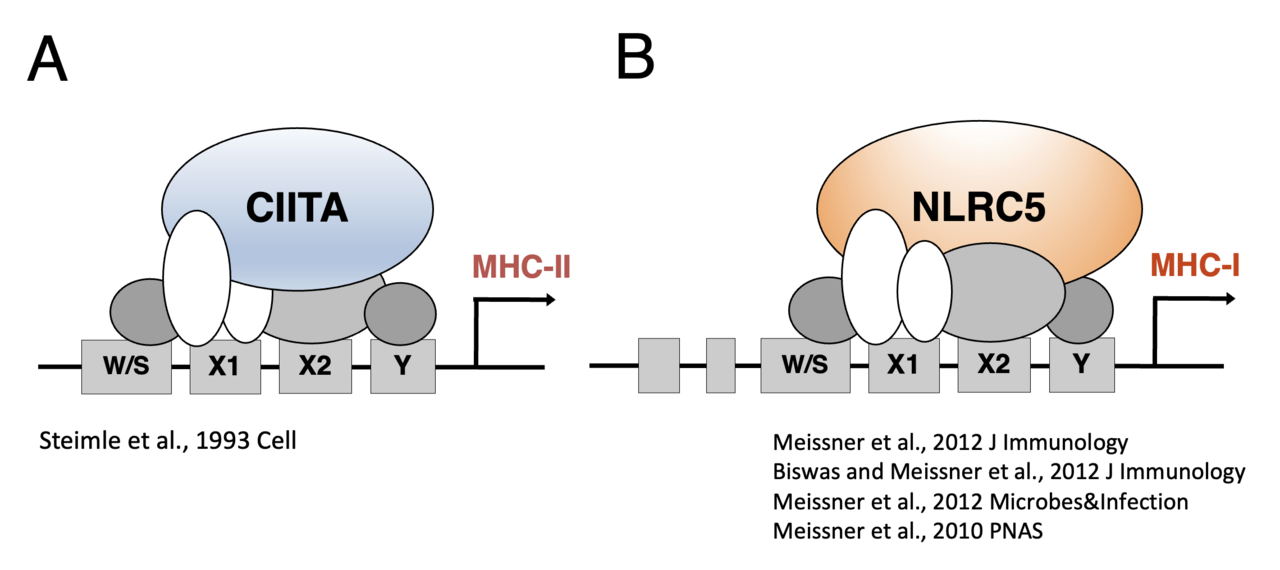
The Human Leukocyte Antigen (HLA) molecules form the immunological fingerprint of a cell and determine if a transplant is being rejected by the recipient's immune system. We hope by understanding how HLA molecules are regulated on the transcriptional level, we will be able to control their expression and mitigate rejection of cells used in cell therapy.
The Immunological Miracle of Pregnancy
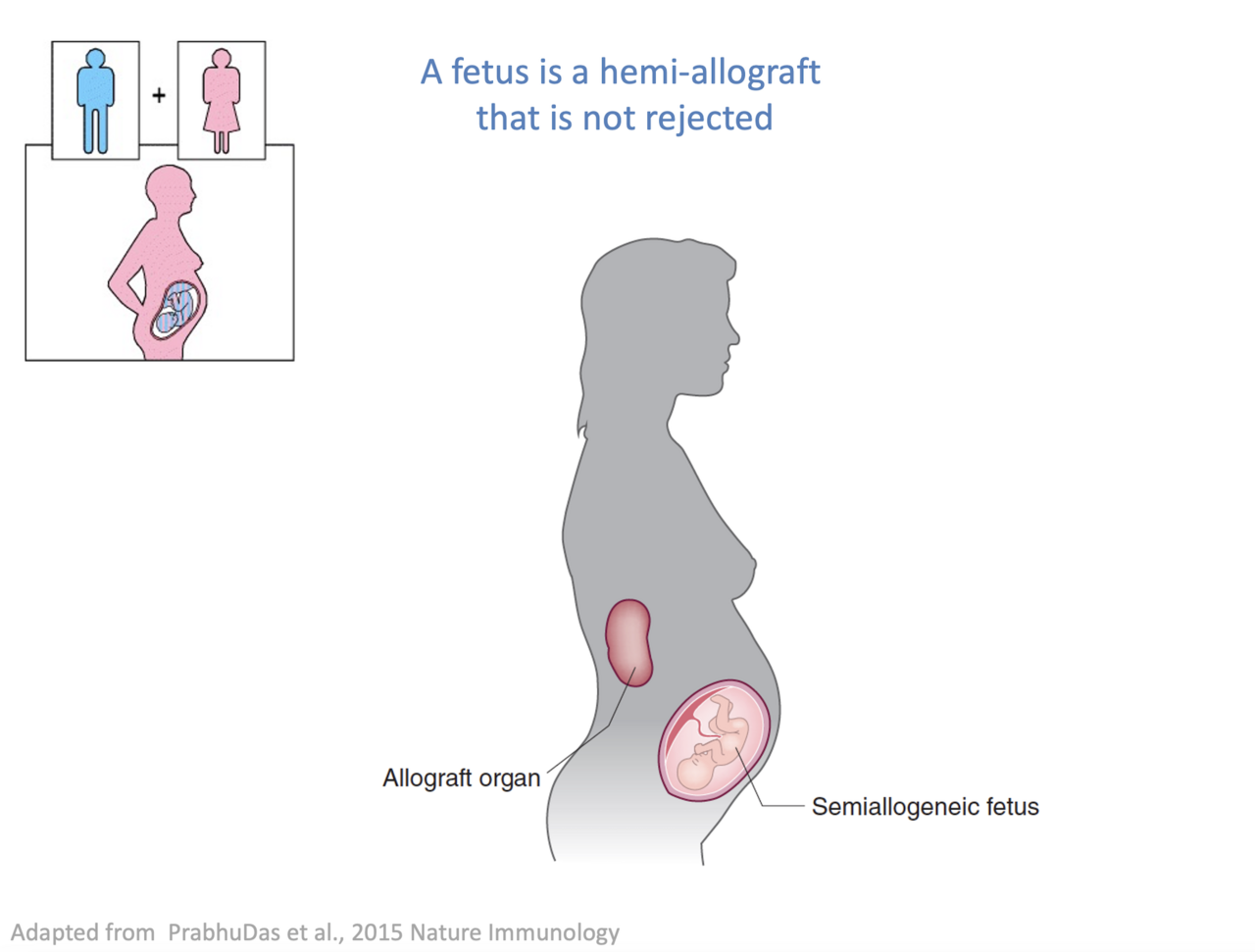
We still don't understand why a baby, which is only 50% identical with the mother, is not being rejected by the maternal immune system. Tolerance to the developing fetus is not systemic since a kidney transplant from the father would readily be rejected. We hope that by understanding how tolerance is established during pregnancy that we can leverage those mechanisms to prevent cell and organ transplants from immune rejection and thereby allowing a broader pool of patients to benefit from emerging cell therapies.
Universal Cells
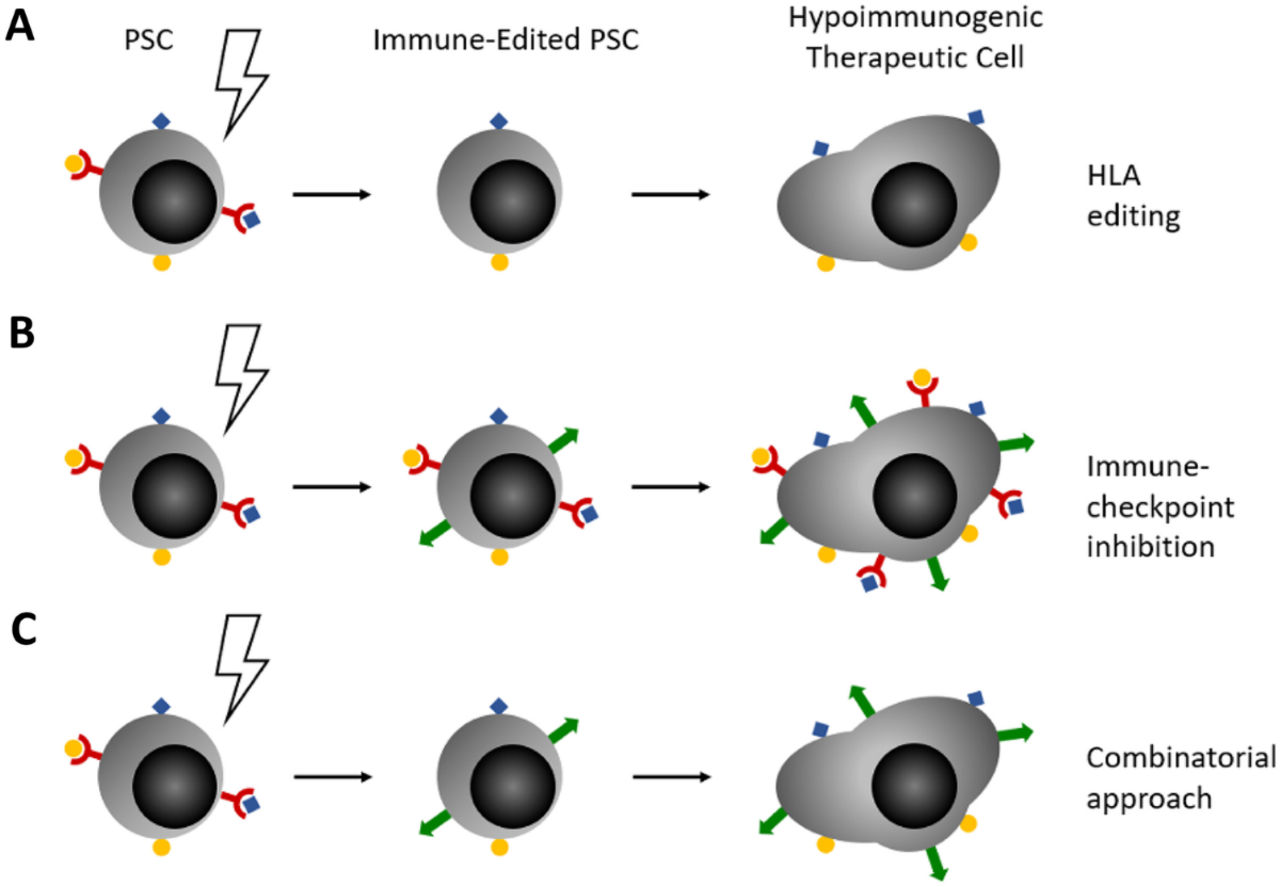
Curr Stem Cell Rep. 2022;8(4):206-218.
We employ cutting edge genome editing technologies to change the immunological fingerprint of induced pluripotent stem cells (iPSC). Such 'universal donor stem cells' can be differentiated into relevant therapeutic cell types and thus democratize access to cell therapy for everyone.
