Research
Alzheimer's disease whole genome sequencing functional interpretation
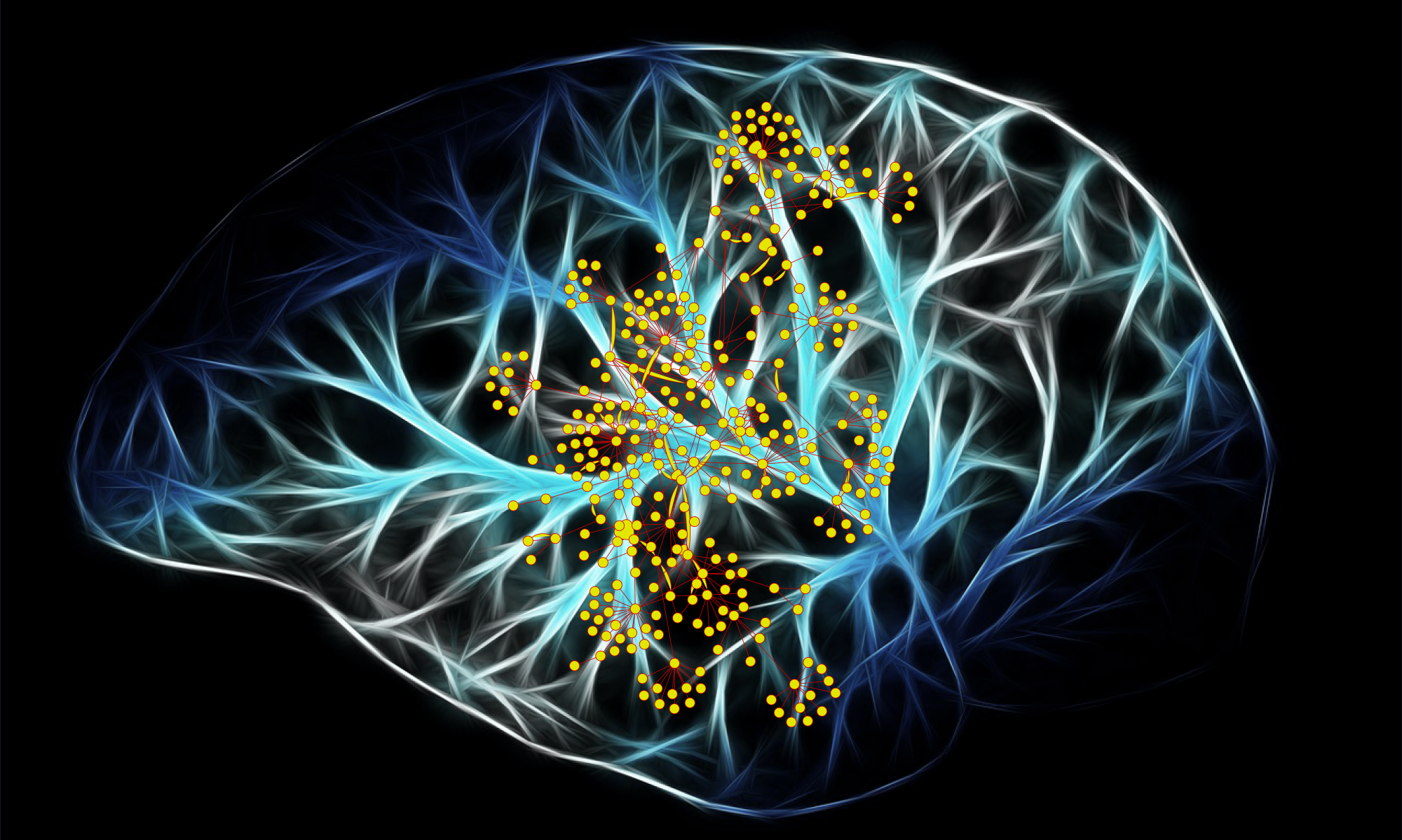
Alzheimer's disease whole genome sequencing functional interpretation
Together with Rudi Tanzi at Massachusetts General Hospital we are working in the Cure Alzheimer's Genome Project to interpret the whole genome sequences of AD afflicted individuals, some in families. We're excited to link these data and to work with our collaborators in findings from the Accelerating Medicines Project - Alzheimer's Disease - enabling functional association with genomic variants. Through recent additional support from the Cure Alzheimer's Foundation, we have completed whole genome sequencing an additional 4000 afflicted AD subjects including methylation studies upon a subset. Rare directional variants from these cohorts are driving our understanding of genetic protection against Alzheimer's Disease pathology.
Mapping Genes to Pathways to Drugs and noncoding RNA
We use systems biology - networks of correlated pathway activities - to connect disease-associated gene activity in pathways to the response of pathways to drug interventions. We use this map to predict those drugs we think would be useful. We published this approach recently going after and successfully addressing sepsis in a mouse model. Now we are building noncoding RNA into our systems as drugs - highly specific - highly efficient with low side effects.
Resilience against Alzheimer's
 Resilience against Alzheimer's
Resilience against Alzheimer's
Because there are susceptible aged individuals who do not show any signs of Alzheimer's, yet may have pathological hallmarks of Alzheimer's - we are particularly interested in finding the basis of resilience against onset of Alzheimer's. Funded by the National Institutes of Health, our project The Alzheimer's Disease Resiliome: Pathway Analysis and Drug Discovery seeks to determine the key pathways that are differentially activated in resilient subjects, and, using drugs and miRNAs, emulate the activation of these pathways in a 3D organoid model of Alzheimer's.
Human Experimental Models
A major problem in translating human genomic variation into targetable therapeutics is that to test targets, we have to use animal models. Animal cures are not human cures and this xenobiotic loss in translation can be a show stopper. So instead we turn the process on its head. We make molecular signatures from post mortem patient tissues. Then we look to see if genes interacting in and around these signatures turn up in human genetic studies. If they do, we consider the signatures associated with the disease at some level.
This means not only thinking about genes that are involved, but how they are interacting to contribute to onset and aetiology of AD. It's a completely different kind of approach to the problem which explores the complex relationships amongst genomic variants and disease pathology. Our current approaches integrate the heterogenous sources of high throughput assays while attempting a synthesis that reflects the functional interactions of genes in pathways.
Industry
The lab has worked closely with industrial partners including Biogen Inc. (disclosure, Dr. Hide used to consult for Biogen) to help analyse and build models of neurodegenerative disease.
Translation of model systems to human health
Using an in silico approach that compares functional activity between model systems such as human iPSC, 3D organoids, mouse and humans, we isolate key genes and pathways in the models that may be contributing to diseases in human subjects. We want to determine the dynamics of pathways as they alter their activity within disease progression, and to predict which subjects will respond to a drug treatment - and predict the drugs they will respond to.
Finding the miRNAs that protect us from Tuberculsosis progression
Mycobacterium tuberculosis infects many people worldwide, and yet only some of them become ill. Together with our collaborator, Reto Guler at the University of Cape Town, we are investigating the genome sequence and miRNAs that regulate the process of infection of macrophages from uninfected subjects who have give us blood samples in Cape Town and also in Kampala. We are building systems models that will allow us to predict effective miRNAs / drugs for treatments in African subjects.
Centenarians - finding the super in super agers

Together with Rudy Tanzi, Doo Kim and Tom Perls, we are performing a multicenter study to understand the phenomenon of super-ager resilience. These individuals, even though they are centenarians, exhibit the cognitive abilities of 70 year olds. Some have significant AD related pathology but little loss of cognitive ability. We are working with the teams to assess neuropsychological, biomarker, neuroimaging and neuropathological phenotyping together with molecular assay of miRNA in blood, brain, and Exome sequencing with whole RNA sequencing of specific regions of the brain normally impacted with pathology. In addition, we are performing spatial transcriptomics, and single nucleus RNA sequencing to determine localized molecular events.
Adult Hippocampal Neurogenesis
One of the fundamental challenges in understanding Alzheimer’s disease (AD) lies in its multifaceted nature—it’s not just about amyloid plaques and tau tangles but about how these pathologies interact with underlying cellular mechanisms. In particular, adult hippocampal neurogenesis (AHN)—the birth of new neurons—emerges as a key player. Surprisingly, resilient individuals, those with AD pathology but no cognitive decline, maintain better AHN, suggesting a potential target for therapeutic strategies.
Instead of simply observing these changes, our approach turns the question on its head: How can AHN and brain-derived neurotrophic factor (BDNF) be harnessed to build cognitive resilience? By combining advanced single-nucleus RNA sequencing, genetic manipulations, and behavioral studies, we’re mapping how AHN interacts with BDNF to maintain hippocampal function. Early findings in mouse models show that increasing AHN alone is insufficient, but when paired with BDNF, cognitive functions improve even amidst pathological conditions.
This approach isn’t just about understanding AD—it’s about translating this knowledge into actionable strategies. By identifying key regulators of AHN and validating their therapeutic potential in human-derived models and pharmacological screens, we aim to bridge the gap between discovery and treatment. It’s not only about which genes are involved but how they work together to preserve cognition. This synthesis of genomics, neurobiology, and resilience modeling could redefine how we think about combating neurodegenerative diseases.
