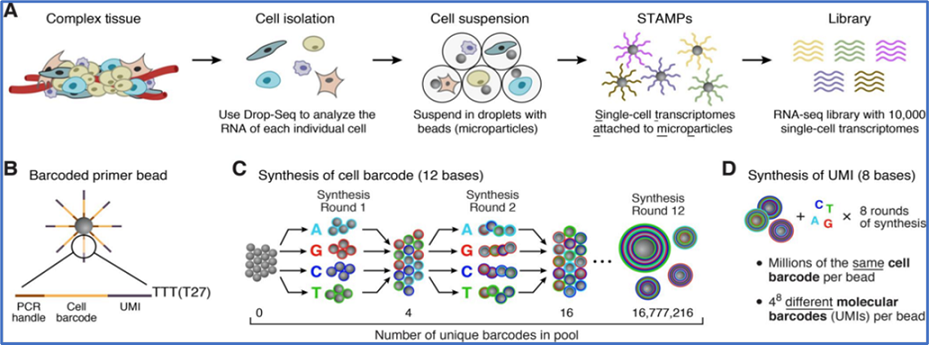
Modern transgenic “tools” enable specific neuron populations to be mapped, monitored, and manipulated in vivo. While these approaches represent an enormous advance, their application requires knowledge of specific transcriptional markers for distinguishing the neurons of interest from their neighbors. Single cell RNA-seq (scRNA-seq) provides an unbiased method for classifying neuron types and their transcriptional markers within a heterogenous tissue.
The aim of these studies is to create an atlas of cell types in the PMC-LC and PAGVL regions. Follow-up studies will identify subpopulations that form the bladder-controlling neurocircuit and that respond to relevant physiological stimuli. Our results will guide strategies to genetically access and manipulate these discrete, functionally-important neuron subpopulations. [pdf of methodology]